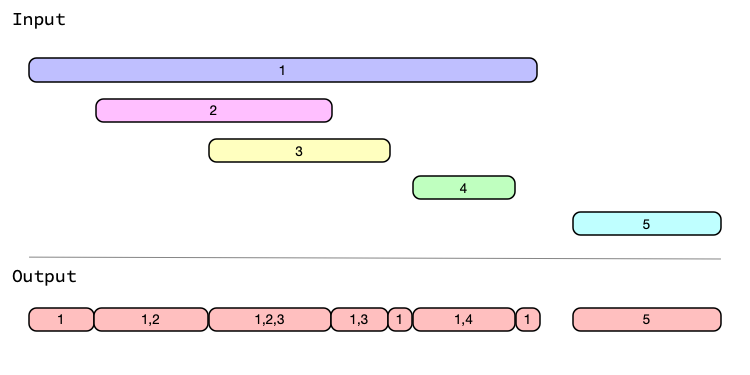
This family of functions revolves around splitting an iv on its endpoints, which results in a new iv that is entirely disjoint (i.e. non-overlapping). The intervals in the resulting iv are known as "splits".
iv_splits()computes the disjoint splits forx.iv_identify_splits()identifies the splits that correspond to each interval inx. It replacesxwith a list of the same size where each element of the list contains the splits that the corresponding interval inxoverlaps. This is particularly useful alongsidetidyr::unnest().iv_locate_splits()returns a two column data frame with akeycolumn containing the result ofiv_splits()and aloclist-column containing integer vectors that map each interval inxto the splits that it overlaps.
Usage
iv_splits(x, ..., on = NULL)
iv_identify_splits(x, ..., on = NULL)
iv_locate_splits(x, ..., on = NULL)Arguments
- x
[iv]An interval vector.
- ...
These dots are for future extensions and must be empty.
- on
[vector / NULL]An optional vector of additional values to split on.
This should have the same type as
iv_start(x).
Value
For
iv_splits(), an iv with the same type asx.For
iv_identify_splits(), a list-of containing ivs with the same size asx.For
iv_locate_splits(), a two column data frame with akeycolumn of the same type asxandloclist-column containing integer vectors.
Examples
library(tidyr)
library(dplyr)
# Guests to a party and their arrival/departure times
guests <- tibble(
arrive = as.POSIXct(
c("2008-05-20 19:30:00", "2008-05-20 20:10:00", "2008-05-20 22:15:00"),
tz = "UTC"
),
depart = as.POSIXct(
c("2008-05-20 23:00:00", "2008-05-21 00:00:00", "2008-05-21 00:30:00"),
tz = "UTC"
),
name = list(
c("Mary", "Harry"),
c("Diana", "Susan"),
"Peter"
)
)
guests <- unnest(guests, name) %>%
mutate(iv = iv(arrive, depart), .keep = "unused")
guests
#> # A tibble: 5 × 2
#> name iv
#> <chr> <iv<dttm>>
#> 1 Mary [2008-05-20 19:30:00, 2008-05-20 23:00:00)
#> 2 Harry [2008-05-20 19:30:00, 2008-05-20 23:00:00)
#> 3 Diana [2008-05-20 20:10:00, 2008-05-21 00:00:00)
#> 4 Susan [2008-05-20 20:10:00, 2008-05-21 00:00:00)
#> 5 Peter [2008-05-20 22:15:00, 2008-05-21 00:30:00)
# You can determine the disjoint intervals at which people
# arrived/departed with `iv_splits()`
iv_splits(guests$iv)
#> <iv<datetime<UTC>>[5]>
#> [1] [2008-05-20 19:30:00, 2008-05-20 20:10:00)
#> [2] [2008-05-20 20:10:00, 2008-05-20 22:15:00)
#> [3] [2008-05-20 22:15:00, 2008-05-20 23:00:00)
#> [4] [2008-05-20 23:00:00, 2008-05-21 00:00:00)
#> [5] [2008-05-21 00:00:00, 2008-05-21 00:30:00)
# Say you'd like to determine who was at the party at any given time
# throughout the night
guests <- mutate(guests, splits = iv_identify_splits(iv))
guests
#> # A tibble: 5 × 3
#> name iv splits
#> <chr> <iv<dttm>> <list<iv<dttm>>>
#> 1 Mary [2008-05-20 19:30:00, 2008-05-20 23:00:00) [3]
#> 2 Harry [2008-05-20 19:30:00, 2008-05-20 23:00:00) [3]
#> 3 Diana [2008-05-20 20:10:00, 2008-05-21 00:00:00) [3]
#> 4 Susan [2008-05-20 20:10:00, 2008-05-21 00:00:00) [3]
#> 5 Peter [2008-05-20 22:15:00, 2008-05-21 00:30:00) [3]
# Unnest the splits to generate disjoint intervals for each guest
guests <- guests %>%
unnest(splits) %>%
select(name, splits)
guests
#> # A tibble: 15 × 2
#> name splits
#> <chr> <iv<dttm>>
#> 1 Mary [2008-05-20 19:30:00, 2008-05-20 20:10:00)
#> 2 Mary [2008-05-20 20:10:00, 2008-05-20 22:15:00)
#> 3 Mary [2008-05-20 22:15:00, 2008-05-20 23:00:00)
#> 4 Harry [2008-05-20 19:30:00, 2008-05-20 20:10:00)
#> 5 Harry [2008-05-20 20:10:00, 2008-05-20 22:15:00)
#> 6 Harry [2008-05-20 22:15:00, 2008-05-20 23:00:00)
#> 7 Diana [2008-05-20 20:10:00, 2008-05-20 22:15:00)
#> 8 Diana [2008-05-20 22:15:00, 2008-05-20 23:00:00)
#> 9 Diana [2008-05-20 23:00:00, 2008-05-21 00:00:00)
#> 10 Susan [2008-05-20 20:10:00, 2008-05-20 22:15:00)
#> 11 Susan [2008-05-20 22:15:00, 2008-05-20 23:00:00)
#> 12 Susan [2008-05-20 23:00:00, 2008-05-21 00:00:00)
#> 13 Peter [2008-05-20 22:15:00, 2008-05-20 23:00:00)
#> 14 Peter [2008-05-20 23:00:00, 2008-05-21 00:00:00)
#> 15 Peter [2008-05-21 00:00:00, 2008-05-21 00:30:00)
# Tabulate who was there at any given time
guests %>%
summarise(n = n(), who = list(name), .by = splits)
#> # A tibble: 5 × 3
#> splits n who
#> <iv<dttm>> <int> <list>
#> 1 [2008-05-20 19:30:00, 2008-05-20 20:10:00) 2 <chr [2]>
#> 2 [2008-05-20 20:10:00, 2008-05-20 22:15:00) 4 <chr [4]>
#> 3 [2008-05-20 22:15:00, 2008-05-20 23:00:00) 5 <chr [5]>
#> 4 [2008-05-20 23:00:00, 2008-05-21 00:00:00) 3 <chr [3]>
#> 5 [2008-05-21 00:00:00, 2008-05-21 00:30:00) 1 <chr [1]>
# ---------------------------------------------------------------------------
x <- iv_pairs(c(1, 5), c(4, 9), c(12, 15))
x
#> <iv<double>[3]>
#> [1] [1, 5) [4, 9) [12, 15)
# You can provide additional singular values to split on with `on`
iv_splits(x, on = c(2, 13))
#> <iv<double>[6]>
#> [1] [1, 2) [2, 4) [4, 5) [5, 9) [12, 13) [13, 15)